Bioinformatics Training Program
We provide bioinformatics training through short workshops and in-depth courses.
Workshops
Next Generation Sequencing Introduction Series:
Workshops on RNA-seq, ChIP-seq to introduce basic concepts of Next-Generation Sequencing (NGS) analysis. The goal of these workshops are to enable researchers to design their studies appropriately and perform preliminary data analyses.
Basic Bioinformatics Skills Series:
Topics include R, data visualization using R, Python programming, Unix and high-performance computing (duration varies from half a day to three days).
In-Depth Next Generation Sequencing Analysis Courses
These intensive courses run for 10-12 days and are aimed at bench biologists interested in learning how to perform NGS data analysis independently using best practices. Topics covered in this course include:
UNIX & High-Performance Computing (Orchestra with HMS-RC) NGS data analysis (RNA-seq, ChIP-seq, Variant calling) Statistical analysis using R Functional analysis downstream of sequence analysis
No prior NGS or command line expertise is required for our workshops or courses unless explicitly stated.
| Date | Title | Lab |
|---|---|---|
| April 3-4, 2017 | Introduction to R | HSCI, HMS, HNDC |
| March 6-7, 2017 | Introduction to UNIX and Orchestra with HMS-RC | HSCI |
| May 31-July 7, 2017 | In-depth NGS Analysis Course | HSCI, HMS, HNDC |
All teaching materials are available online on Github.
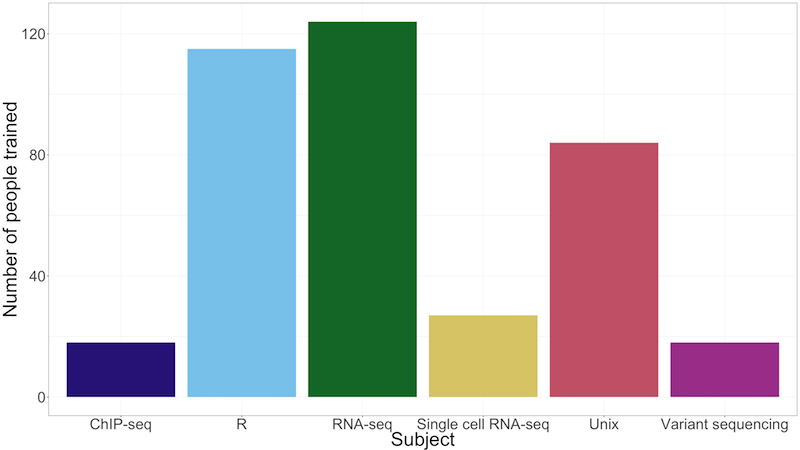
Statistics
Team

Radhika Khetani
Lead Trainer

Meeta Mistry
Research Associate

Mary Piper
Research Associate
